Resolved Vector Velocities#
Read in and plot the CEDAR resolved vector velocity “vvels” data product.
%matplotlib inline
import datetime
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
import re
import h5py
import numpy as np
import matplotlib.gridspec as gridspec
import madrigalWeb.madrigalWeb
import os
# Download the file that we need to run these examples
vfilepath= '../data/pfa20240108.001_lp_vvels_01min.001.h5'
if not os.path.exists(vfilepath):
madrigalUrl='http://cedar.openmadrigal.org'
data = madrigalWeb.madrigalWeb.MadrigalData(madrigalUrl)
user_fullname = 'Student Example'
user_email = 'isr.summer.school@gmail.com'
user_affiliation= 'ISR Summer School 2020'
url='/opt/cedar3/experiments4/2024/pfa/08jan24a/pfa20240108.001_lp_vvels_01min.001.h5'
print('Downloading data file...')
file = data.downloadFile(url, vfilepath,
user_fullname, user_email, user_affiliation,'hdf5')
print('...Done!')
Downloading data file...
...Done!
Line-of-Sight Velocity [ms-1]#
The velocity array is in the 2D Parameters:
VIPN: perp north
VIPE: perp east
VI6: antiparallel
Dimensons: Nlatitude X Nrecords
with h5py.File(vfilepath, 'r') as v:
times=[datetime.datetime(1970,1,1)+datetime.timedelta(seconds=int(t)) for t in v['Data']['Array Layout']['timestamps']]
cgm_lat=np.array(v['Data']['Array Layout']['cgm_lat'])
vipn = np.array(v['Data']['Array Layout']['2D Parameters']['vipn'])
dvipn = np.array(v['Data']['Array Layout']['2D Parameters']['dvipn'])
vipe = np.array(v['Data']['Array Layout']['2D Parameters']['vipe'])
dvipe = np.array(v['Data']['Array Layout']['2D Parameters']['dvipe'])
vi6 = np.array(v['Data']['Array Layout']['2D Parameters']['vi6'])
dvi6 = np.array(v['Data']['Array Layout']['2D Parameters']['dvi6'])
---------------------------------------------------------------------------
OSError Traceback (most recent call last)
Cell In[3], line 1
----> 1 with h5py.File(vfilepath, 'r') as v:
2 times=[datetime.datetime(1970,1,1)+datetime.timedelta(seconds=int(t)) for t in v['Data']['Array Layout']['timestamps']]
3 cgm_lat=np.array(v['Data']['Array Layout']['cgm_lat'])
File /usr/share/miniconda/envs/buildjupyterbook/lib/python3.10/site-packages/h5py/_hl/files.py:564, in File.__init__(self, name, mode, driver, libver, userblock_size, swmr, rdcc_nslots, rdcc_nbytes, rdcc_w0, track_order, fs_strategy, fs_persist, fs_threshold, fs_page_size, page_buf_size, min_meta_keep, min_raw_keep, locking, alignment_threshold, alignment_interval, meta_block_size, **kwds)
555 fapl = make_fapl(driver, libver, rdcc_nslots, rdcc_nbytes, rdcc_w0,
556 locking, page_buf_size, min_meta_keep, min_raw_keep,
557 alignment_threshold=alignment_threshold,
558 alignment_interval=alignment_interval,
559 meta_block_size=meta_block_size,
560 **kwds)
561 fcpl = make_fcpl(track_order=track_order, fs_strategy=fs_strategy,
562 fs_persist=fs_persist, fs_threshold=fs_threshold,
563 fs_page_size=fs_page_size)
--> 564 fid = make_fid(name, mode, userblock_size, fapl, fcpl, swmr=swmr)
566 if isinstance(libver, tuple):
567 self._libver = libver
File /usr/share/miniconda/envs/buildjupyterbook/lib/python3.10/site-packages/h5py/_hl/files.py:238, in make_fid(name, mode, userblock_size, fapl, fcpl, swmr)
236 if swmr and swmr_support:
237 flags |= h5f.ACC_SWMR_READ
--> 238 fid = h5f.open(name, flags, fapl=fapl)
239 elif mode == 'r+':
240 fid = h5f.open(name, h5f.ACC_RDWR, fapl=fapl)
File h5py/_objects.pyx:54, in h5py._objects.with_phil.wrapper()
File h5py/_objects.pyx:55, in h5py._objects.with_phil.wrapper()
File h5py/h5f.pyx:102, in h5py.h5f.open()
OSError: Unable to synchronously open file (file signature not found)
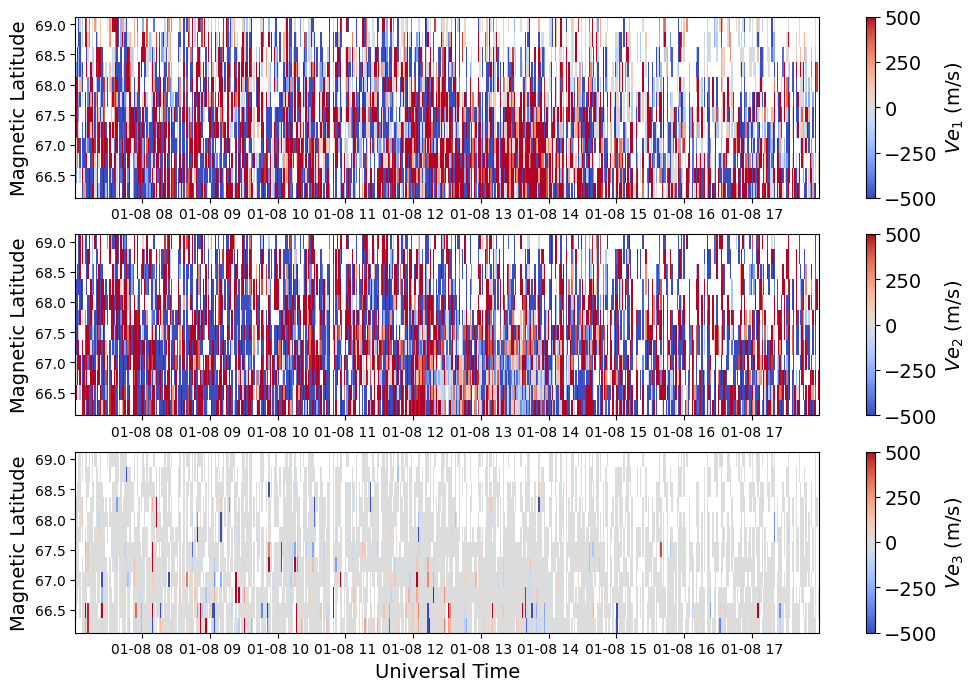
Resolved Vector Velocity Data#
with h5py.File(vfilepath, 'r') as v:
utime= np.array([datetime.datetime(1970,1,1)+datetime.timedelta(seconds=int(t)) for t in v['Data']['Array Layout']['timestamps']])
mlat= np.array(v['Data']['Array Layout']['cgm_lat'])
vipn = np.array(v['Data']['Array Layout']['2D Parameters']['vipn'])
vipe = np.array(v['Data']['Array Layout']['2D Parameters']['vipe'])
vi6 = np.array(v['Data']['Array Layout']['2D Parameters']['vi6'])
#print(np.array(v['Data']['Array Layout']['2D Parameters']['Data Parameters']))
vel = np.array([vipe,vipn,vi6]).T
time = utime.astype('datetime64[s]')
plt.rcParams.update({'font.size': 14})
fig = plt.figure(figsize=(12,8))
gs = gridspec.GridSpec(3,1)
ax = [fig.add_subplot(gs[i]) for i in range(3)]
for i in range(3):
c = ax[i].pcolormesh(time, mlat, vel[:,:,i].T, vmin=-500., vmax=500., cmap='coolwarm')
ax[i].set_ylabel('Magnetic Latitude', fontsize=14)
ax[i].tick_params(axis='both', labelsize=10) # Change tick label size for both axes
fig.colorbar(c, label=fr'$Ve_{i+1}$ (m/s)')
ax[2].set_xlabel('Universal Time')
Text(0.5, 0, 'Universal Time')
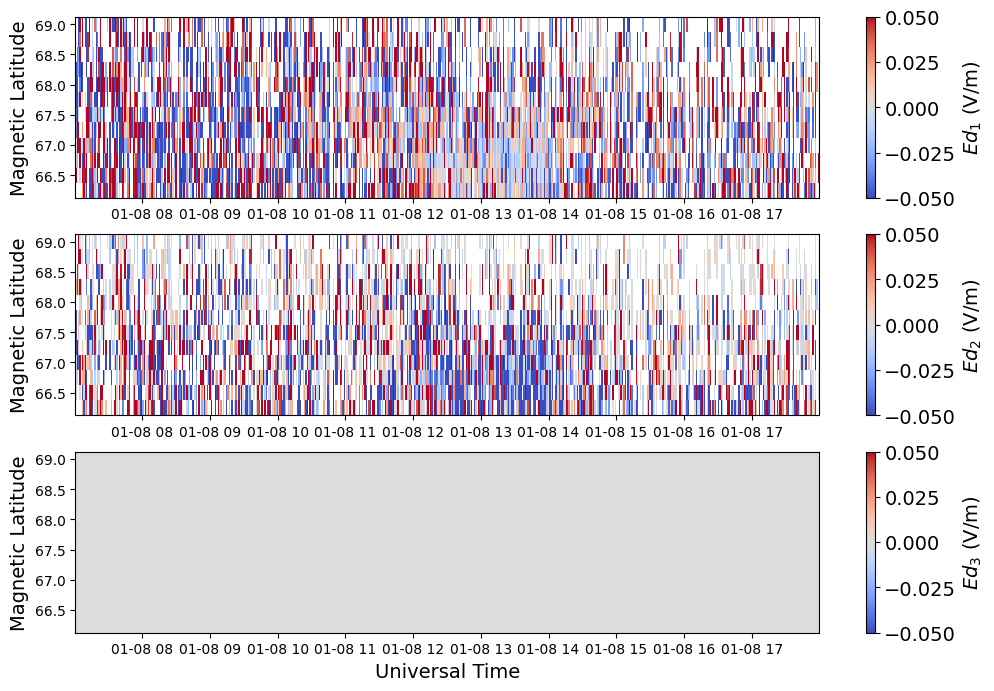
Electric Field#
The electric field and velocity in the resolved velocities files are not independent measurements. Both are provided for user convenience. By definition, there is no parallel electric field component
with h5py.File(vfilepath, 'r') as v:
param = list(v['Data']['Array Layout']['2D Parameters']['Data Parameters'])
utime= np.array([datetime.datetime(1970,1,1)+datetime.timedelta(seconds=int(t)) for t in v['Data']['Array Layout']['timestamps']])
mlat= np.array(v['Data']['Array Layout']['cgm_lat'])
epn = np.array(v['Data']['Array Layout']['2D Parameters']['epn'])
epe = np.array(v['Data']['Array Layout']['2D Parameters']['epe'])
e3 = np.zeros_like(epe)
efield = np.array([epe,epn,e3]).T
time = utime.astype('datetime64[s]')
plt.rcParams.update({'font.size': 14})
fig = plt.figure(figsize=(12,8))
gs = gridspec.GridSpec(3,1)
ax = [fig.add_subplot(gs[i]) for i in range(3)]
for i in range(3):
c = ax[i].pcolormesh(time, mlat, efield[:,:,i].T, vmin=-0.05, vmax=0.05, cmap='coolwarm')
ax[i].set_ylabel('Magnetic Latitude')
ax[i].tick_params(axis='both', labelsize=10) # Change tick label size for both axes
fig.colorbar(c, label=fr'$Ed_{i+1}$ (V/m)')
ax[2].set_xlabel('Universal Time')
Text(0.5, 0, 'Universal Time')